Single-cell sequencing: what it is and why researchers in industry and academia want it
- ArgenTag Bioscience
- Jan 23, 2024
- 5 min read
Updated: Aug 28, 2024

Image Credit: Generated with Microsoft Bing AI
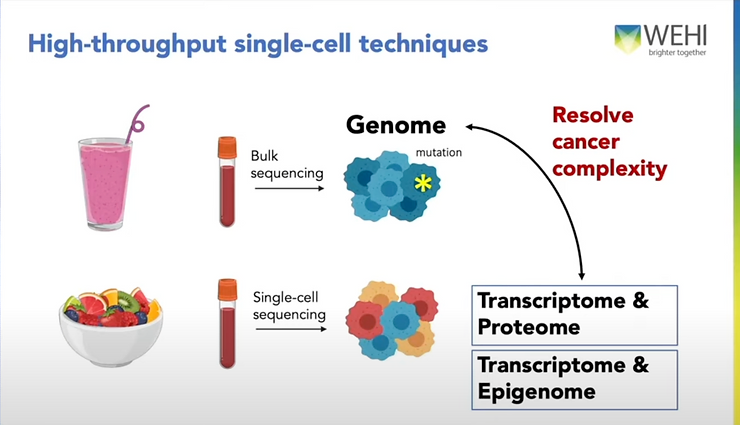
As Mark’s oncologists receive his leukemia cell sequencing results to design a targeted treatment, they get a general understanding of the initial genetic variations and activity of his genes. However, the traditional sequencing methods they used are only an average of all the immune cells in Mark’s blood or bone marrow, which means they cannot attribute specific mutations to specific immune cell populations. It’s like seeing the fog of a fire from afar without being able to point to the source. Even when using flow cytometry to sort and isolate specific types of immune cells, Mark’s doctors find themselves limited in the number of genes they can analyze, which isn’t of much help considering leukemia’s high genetic heterogeneity.
Though hypothetical, this is far from being an isolated case. Many diseases that present similar symptoms may respond differently to different treatments depending on what type of immune cell the disease is stemming from. Other patients may struggle with ovarian, breast, lung, or other cancers and immune diseases that require a precise, cell-level understanding of the patient’s genetics. The problem is that most tissues, including blood, are like a smoothie: a multitude of cells mixed together that when sequenced together, only offer an average taste, making any treatment a shot in the dark.
Smoothie analogy. Image credits to Rachel Thijssen at the Oxford Nanopore conference
Single-cell sequencing is the solution that does just what it sounds like: help researchers know the sequences of each individual cell in a sample separately, not as an average of the others.
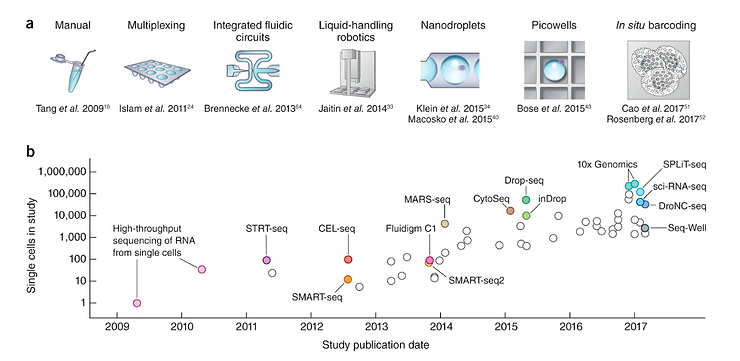
If this sounds tedious or impossible, it’s because it used to be that way. For Mark’s and millions of other patients’ fortune, recent advances in microfluidics, optics and AI have led to progress akin to Moore’s Law in the number of cells that researchers can sequence individually at a time, as well as the speed and at which they can do it. Over the last couple of decades, the world has gone from heavily manual methods that could sequence less than 100 individual cells at a time, to more efficient ones that are close to reaching 1 million cells sequenced from a single sample.
Scaling of sc-RNAseq experiments from 2009 to 2017. Image credits to Svensson et.al
Beyond the aforementioned examples, single-cell sequencing has allowed developmental biologists to understand how organ development processes can result in malformations and disease, microbiologists to track how our gut microbiome may modify the drugs we take, and epidemiologists to identify different cell types in the nose that could explain COVID-19 high transmission rates. Our favorite example is perhaps gene identification in sickle cell disease therapy, which just in 2023 became the very first disease to get a gene therapy treatment approved by the FDA.
“It’s like a flight recorder, where you are watching what went wrong and not just looking at a snapshot at the end” Jonathan Weissman, stem cell biologist at UCSF, in Science
Who uses single-cell and why?
Such advancements haven’t gone unnoticed by the new wave of big-tech-funded biotech research organizations. The Chan-Zuckerberg Initiative (CZI) has so far deployed more than $200 million USD in funding towards single-cell projects and in October of 2023, they announced that their Single-Cell Biology team in New York will use single-cell sequencing and AI to unlock the vast information stored in immune cells to monitor and manage organ and tissue health. CZI is also a contributor to the Human Cell Atlas consortium which is a global collaboration even larger than the Human Genome Project that aims to transform our understanding of the 37.2 trillion cells in the human body by using single-cell genomics to create comprehensive reference maps of each type of cell.
Image credit: Mark Zuckerberg & Dr. Priscilla Chan: Curing All Human Diseases & the Future of Health & Technology
Around the world, biotech startups such as Celsius Therapeutics and Modulo Bio have single-cell genomics as their main differentiator to develop precision therapies for complex diseases. Given that some cell populations may be present in some patients but not in others, sequencing individual cells makes it possible to understand the key differences between responders and non-responders to therapy and thus create patient subsets for drug development. Single-cell sequencing also proves helpful in undergoing precise CRISPR gene editing analyses when different types of cells are cultured together to mimic the body environment or, similarly, when validating organoid models through sequencing.
Others such as Phenomic AI are using single-cell sequencing to understand how cancer cells hijack the fibroblasts that surround them and how these hijacked fibroblasts then shut down T-cells that are usually programmed to attack the cancer cells. Similar to Celsius and Modulo, they can now get a clearer understanding of drug target expression across a broad range of tissues to develop truly specific and highly potent therapeutics.
Tumor with cancer and cancer-associated fibroblasts. Image credits to Phenomic AI
Limitations and opportunities
All of these amazing developments make it clear that single-cell technology has become the new paradigm in disease understanding and therapeutic development. Today, however, the crucial step of barcoding each cell’s genetic material remains like a patch that requires the use of highly specialized equipment like microfluidic devices offered by companies like 10x genomics, plus the use of short-read sequencing devices offered by companies like Illumina. Knowing that the average human gene length is around 20 thousand base pairs, existing solutions are further limiting researchers studying gene isoforms present in complex diseases (more on that in our next blog post).
"Long-read sequencing technologies are the next frontier in single-cell genomics at scale" Conte et.al, Human Technopole in Cell
ArgenTag: the next gen in single-cell sequencing
At ArgenTag, we are developing the first instrument-free, high-resolution, cost-effective kit that makes single-cell, long-read sequencing available to anyone, anywhere.
Picture attempting to comprehend a movie by watching just a few minutes from different scenes. It's quite a challenge, isn't it? This mirrors the limitations of traditional sequencing methods, which provide only fragmented glimpses. Now, envision being able to capture entire scenes in one go. That's precisely what ArgenTag technology aims to achieve. By enabling longer sequences, it allows us to grasp the complete narrative, gain clearer insights, eliminate guesswork, and achieve a more profound understanding of the cellular story. With this revolutionary technology, we attain a holistic perspective of a cell's genome or transcriptome. It uncovers intricate details—such as isoforms, alternative genetic pathways, and unique cell changes—that were previously concealed in the shadows.
We’re working with researchers at Mt Sinai, Weill Cornell, NYU Langone Health and a dozen of biotech companies to unlock the next wave of discoveries. If this sounds of interest to you too, we would love to get in touch!
In our next post, we will go deeper into the current demand and opportunities in long-read sequencing. Stay tuned for that.
By Sofia Sanchez